Diversity of Metabolism in Procaryotes (page 2)
(This chapter has 8 pages)
© Kenneth Todar, PhD
NAD
Another coenzyme commonly involved in energy-producing metabolism,
derived
from the vitamin niacin, is the pyridine nucleotide, NAD (Nicotinamide
Adenine Dinucleotide). The basis for chemical transformations of
energy
usually involves oxidation/reduction reactions. For a biochemical to
become
oxidized, electrons must be removed by an oxidizing agent. The
oxidizing
agent is an electron acceptor that becomes reduced in the reaction.
During
the reaction, the oxidizing agent is converted to a reducing agent that
can add its electrons to another chemical, thereby reducing it, and
reoxidizing
itself. The molecule that usually functions as the electron carrier in
these types of coupled oxidation-reduction reactions in
biological
systems is NAD and its phosphorylated derivative,
NADP. NAD
or NADP can become alternately oxidized or reduced by the loss or gain
of two electrons. The oxidized form of NAD is symbolized NAD; the
reduced
form is symbolized as NADH, NADH2 or NADH + H+.
The structure of NAD is drawn below.

Figure 3. The Structure of
NAD.
(a) Nicotinamide Adenine Dinucleotide is composed of two nucleotide
molecules:
Adenosine monophosphate (adenine plus ribose-phosphate) and
nicotinamide
ribotide (nicotinamide plus ribose-phosphate). NADP has an identical
structure
except that it contains an additional phosphate group attached to one
of
the ribose residues. (b) The oxidized and reduced forms of of the
nicotinamide
moiety of NAD. Nicotinamide is the active part of the molecule where
the
reversible oxidation and reduction takes place. The oxidized form of
NAD
has one hydrogen atom less than the reduced form and, in addition, has
a positive charge on the nitrogen atom which allows it to accept a
second
electron upon reduction. Thus the correct way to symbolize the reaction
is NAD+ + 2H----->NADH + H+.
However, for convenience, we will hereafter use the symbols NAD and NADH2.
Many
bacterial
protein toxins including the cholera toxin, pertussis toxin and
diphtheria
toxin, exert their enzymatic activity using NAD as a co-substrate. The
toxins are referred to as ADP-ribosylation toxins, because they cleave
NAD into nicotinamide plus ADP-ribose (ADPR) and then transfer the ADPR
to some host molecule. For example, the diphtheria toxin transfers ADPR
to elongation factor 2, irreversibly inactivating its role in chain
elongation
during protein synthesis. Thus, the biological activity of the
diphtheria
toxin is to inhibit protein synthesis in eucaryotic cells.
Coenzyme A
Coenzyme A is another coenzyme frequently involved in
energy-generating
metabolism of procaryotes. Coenzyme A is involved in a type of
ATP-generating
reaction seen in some fermentative bacteria and in all respiratory
organisms.
The reaction occurs in association with the oxidation of keto acids
such
as pyruvic acid and alpha ketoglutaric acid. These substrates are
central
to glycolysis and the TCA cycle, respectively, and they are direct or
indirect
precursors of several essential macromolecules in a cell. The
oxidations
of pyruvate and alpha ketoglutarate, involving Coenzyme A, NAD, a
dehydrogenation
reaction and a decarboxylation reaction, are two of the most
important,
and complex, reactions in metabolism.
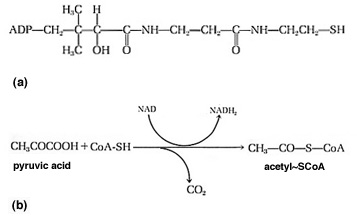
Figure 4. (a) The Structure
of Coenzyme A. CoA-SH is a derivative of ADP. The molecule shown here
attached
to ADP is pantothenic acid, which carries a terminal thiol (-S) group.
(b) the oxidation of the keto acid, pyruvic acid, to acetyl~SCoA. This
is the reaction that enters two carbons from pyruvate into the TCA
cycle.
In the oxidation of keto acids, coenzyme A (CoA or CoASH) becomes
attached
through a thioester linkage (~S) to the carboxyl group of the oxidized
product. Part of the energy released in the oxidation is conserved in
the
thioester bond. This bond energy can be subsequently used to synthesize
ATP, as in the case of the clostridia that convert acetyl~SCoA +ADP +
Pi-------->
acetic acid + CoASH + ATP. Or in the case of respiratory
organisms,
the thioester bond energy is expended when acetyl~SCoA condenses with
oxalacetate
in order to drive the TCA cycle into its oxidative branch.
ATP Synthesis in
Procaryotes
The objective of a catabolic pathway is to make ATP: to transform
either
chemical energy or electromagnetic (light) energy into the chemical
energy
contained within the high-energy bonds of ATP. Cells fundamentally can
produce ATP in two ways: substrate level phosphorylation and electron
transport phosphorylation.
Substrate level phosphorylation (SLP) is The simplest,
oldest
and least-evolved way to make ATP. In a substrate level
phosphorylation,
ATP is made during the conversion of an organic molecule from one form
to another. Energy released during the conversion is partially
conserved
during the synthesis of the high energy bond of ATP. SLP occurs during
fermentations and respiration (the TCA cycle), and even during some
lithotrophic
transformations of inorganic substrates.

Figure 5. Three examples of
substrate level phosphorylation. (a) and (b) are the two substrate
level
phosphorylations that occur during the Embden Meyerhof pathway, but
they
occur in all other fermentation pathways which have an Embden-Meyerhof
component. (c) is a substrate level phosphorylation found in Clostridium
and
Bifidobacterium.
These
are two anaerobic (fermentative) bacteria who learned how to make one
more
ATP from glycolysis beyond the formation of pyruvate.
Electron Transport Phosphorylation (ETP) is a much more
complicated
affair that evolved long after SLP. Electron Transport Phosphorylation
takes place during respiration, photosynthesis, lithotrophy and
possibly
other types of bacterial metabolism. ETP requires that electrons
removed
from substrates be dumped into an electron transport system (ETS)
contained
within a membrane. The electrons are transferred through the ETS to
some
final electron acceptor in the membrane (like O2 in aerobic
respiration) , while their traverse through the ETS results in the
extrusion
of protons and the establishment of a proton motive force (pmf)
across the membrane. An essential component of the membrane for
synthesis
of ATP is a membrane-bound ATPase (ATP synthetase) enzyme. The
ATPase
enzyme transports protons, thereby utilizing the pmf (protons) during
the
synthesis of ATP. The idea in electron transport phosphorylation is to
drive electrons through an ETS in the membrane, establish a pmf, and
use
the pmf to synthesize ATP. Obviously, ETP take a lot more "gear" than
SLP,
in the form of membranes, electron transport systems, ATPase enzymes,
etc.
A familiar example of energy-producing and energy-consuming
functions
of the bacterial membrane, related to the establishment and use of pmf
and the production of ATP, is given in the following drawing of the
plasma
membrane of Escherichia coli.

Figure 6. The plasma membrane
of
Escherichia coli. The membrane in cross-section reveals various
transport systems, the flagellar motor apparatus (S and M rings), the
respiratory
electron transport system, and the membrane-bound ATPase enzyme.
Reduced
NADH + H+ feeds pairs of electrons into the ETS. The ETS is the
sequence
of electron carriers in the membrane [FAD --> FeS --> QH2
(Quinone) -->
(cytochromes) b --> b --> o] that ultimately reduces O2
to H2O
during respiration. At certain points in the electron transport
process,
the electrons pass "coupling sites" and this results in the
translocation
of protons from the inside to the outside of the membrane, thus
establishing
the proton motive force (pmf) on the membrane. The pmf is used in three
ways by the bacterium to do work or conserve energy: active transport
(e.g.
lactose and proline symport; calcium and sodium antiport); motility
(rotation
of the bacterial flagellum), and ATP synthesis (via the ATPase enzyme
during
the process of oxidative phosphorylation or electron transport
phosphorylation).
chapter continued
Previous Page | Next Page